I recently got my Arduino Uno R3 board this week and simply love how easy it is to use! My first muck around project was using a MaxSonar EZ3 sonar sensor to display the range readings to a serial LED segment display.
The MaxSonar’s output of (Vcc/512) per inch was not as accurate as I’d like it to be. I assumed Vcc = 5V. I decided to calibrate it by taking a few readings using a tape measure and reading the raw readings from the 10bit analog pin, hoping to fit a straight line. Plotting this data gave the following graph
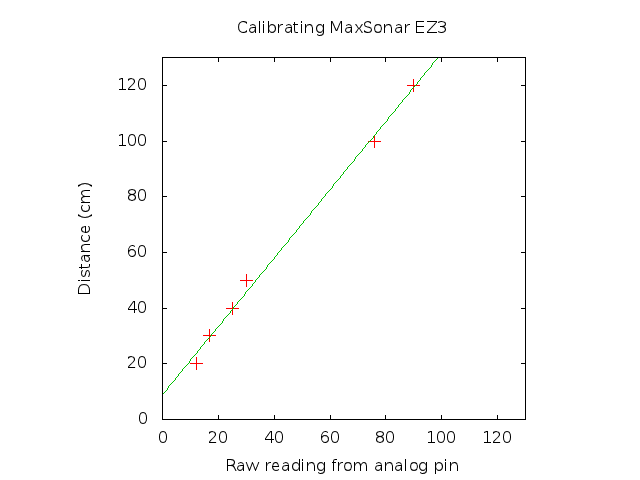 As I hoped it’s a nice linear relationship. I’ve only collected data up to 120cm, ideally you would do it up to the range you’re interested in working to. Fitting a y = mx + c line gave the following values
As I hoped it’s a nice linear relationship. I’ve only collected data up to 120cm, ideally you would do it up to the range you’re interested in working to. Fitting a y = mx + c line gave the following values
m = 1.2275
c = 8.8524
So now I have a nice formula for converting the analog reading to range in centimetres. In practice I use fixed point integer maths.
centimetres = 1.2275*analog_reading + 8.8524
The full sketch up code is
void setup()
{
Serial.begin(9600);
}
int init_loop = 0;
void loop()
{
// NOTE to myself: Arduino Uno int is 16 bit
const int sonarPin = A0;
int dist = analogRead(sonarPin);
// linear equation relating distance vs analog reading
// y = mx + c
// m = 1.2275
// c = 8.8524
// fixed precision scaling by 100
dist = dist*123 + 885;
dist /= 100;
// output the numbers
int thousands = dist / 1000;
int hundreds = (dist - thousands*1000)/100;
int tens = (dist - thousands*1000 - hundreds*100)/10;
int ones = dist - thousands*1000 - hundreds*100 - tens*10;
// Do a few times to make sure the segment display recieves the data ...
if(init_loop < 5) {
// Clear the dots
Serial.print(0x77);
Serial.print(0x00);
// Set brightness
Serial.print("z");
Serial.print(0x00); // maximum brightness
init_loop++;
}
// reset position, needed in case the serial transmission stuffs up
Serial.print("v");
// if thousand digit is zero don't display anything
if(thousands == 0) {
Serial.print("x"); // blank
}
else {
Serial.print(thousands);
}
// if thousand digit and hundred digit are zero don't display anything
if(thousands == 0 && hundreds == 0) {
Serial.print("x");
}
else {
Serial.print(hundreds);
}
// always display the tens and ones
Serial.print(tens);
Serial.print(ones);
delay(100);
}
To get the raw analog reading comment out the bit where it calculates the distance using the linear equation.
Below is a picture of the hardware setup. The MaxSonar is pointing up the ceiling, which I measured with a tape measure to be about 219 cm. The readout from the Arduino is 216cm, so it’s not bad despite calibrating up to 120cm only.
Quirks and issues
It took me a while to realise an int is 16bit on the Arduino! (fail …) I was banging my head wondering why some simple integer maths was failing. I have to be conscious of my scaling factors when doing fixed point maths.
After compiling and uploading code to the Arduino I found myself having to unplug and plug it back in for the serial LED segment display to function correctly. It seems to leave it in an unusable state during the uploading process.